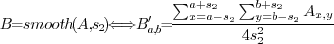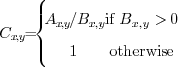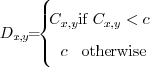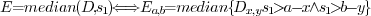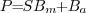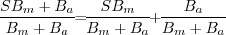| Home | Papers | Reports | Projects | Code Fragments | Dissertations | Presentations | Posters | Proposals | Lectures given | Course notes |
|
|
Adaptive contrast enhancement of two-dimensional electrophoretic protein gel images facilitates visualization, orientation and alignmentWerner Van Belle1* - werner@yellowcouch.org, werner.van.belle@gmail.com Abstract : Two-dimensional polyacrylamide gel electrophoresis (2-DE) is a powerful technique to discriminate post-translationally modified protein isoforms. However, all steps of 2-DE preparation and gel-staining may introduce unwanted artefacts, including inconsistent variation of background intensity over the entire 2-DE gel image. Background intensity variations limit the accuracy of gel orientation, overlay alignment and spot detection methods. We present a compact and efficient denoising algorithm that adaptively enhances the image contrast and then, through thresholding and median filtering, removes the gray-scale range covering the background. Applicability of the algorithm is demonstrated on immuno-blots, isotope labeled gels, and protein stained gels. Validation is performed in contexts of i) automatic gel orientation based on Hough transformation, ii) overlay alignment based on cross correlation and iii) spot detection. In gel-stains with low background variability, e.g. Sypro Ruby, denoising will lower the spot detection sensitivity. In gel regions with high background levels denoising enhances spot detection. We propose that the denoising algorithm prepares images with high background for further automatic analysis, without requiring manual input on a gel-to-gel basis.
Keywords:
adaptive contrast enhancement, 2 dimensional electrophoretic gels, 2D gel image denoising, SDS-PAGE, background estimation hough fourier transform alignment rotation |
Table Of Contents
| Introduction Materials and methods Results and Discussion | Concluding Remarks Acknowledgments Bibliography |
Introduction
Materials and methods
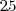 million
NB4 cells by trichloroacetic acid (TCA) precipitation [[13]].
For 2-DE gels with
million
NB4 cells by trichloroacetic acid (TCA) precipitation [[13]].
For 2-DE gels with 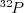 labeled proteins, 10
million MOLM-13
cells were incubated with
labeled proteins, 10
million MOLM-13
cells were incubated with 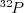 labeled ATP and
thereafter precipitated
in TCA as previously described [[14]]. The AML cell lines
NB4
and MOLM-13 were cultured according to standard procedures
(www.atcc.org).
For all 2-DE experiments, precipitated proteins were diluted in a
rehydration buffer containing 7 M urea, 2 M thiourea, 4%
3-[(3-cholamidopropyl)dimethyl-ammonio]-1-propanesulfonate
(CHAPS), 2% ampholytes and 100 mM dithiothreitol (DTT). The focusing
strips were incubated with protein in rehydration buffer over night
at room temperature.
labeled ATP and
thereafter precipitated
in TCA as previously described [[14]]. The AML cell lines
NB4
and MOLM-13 were cultured according to standard procedures
(www.atcc.org).
For all 2-DE experiments, precipitated proteins were diluted in a
rehydration buffer containing 7 M urea, 2 M thiourea, 4%
3-[(3-cholamidopropyl)dimethyl-ammonio]-1-propanesulfonate
(CHAPS), 2% ampholytes and 100 mM dithiothreitol (DTT). The focusing
strips were incubated with protein in rehydration buffer over night
at room temperature. 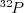 -gels,
IEF on 13 cm 3-10 NL pre-cast focusing strips (GE Healthcare Bio-sciences)
was performed on an IPGphor unit (GE Healthcare Bio-sciences). IEF
conditions used were an initial gradient up to 500 V for 10 minutes,
a second gradient up to 4000 V for 150 minutes, and a final step at
8000 V for 130 minutes. Following IEF, equilibration was performed
in two steps using an equilibration buffer (EQ) containing 6 M urea,
50 mM Tris-HCl, 30% glycerol and 2% SDS. Step one was performed
in EQ containing 1.5% DTT for 12 minutes and step two in EQ containing
4.5% iodoacetamide for 5 minutes. The second dimension was run on
a 10% SDS-polyacrylamide gel. 2-DE gels were stained with Sypro Ruby
dye (Bio-Rad) according to the manufacturer's recommendations. These
gels were imaged either with UV light as an excitation source and
a 670 nm emission filter (35 nm wide band) on a Kodak image station
2000R or with a 457 nm excitation laser source and a 610 nm emission
filter on a Typhoon imager 9400 (GE Healthcare Bio-sciences AB,
Uppsala,
Sweden). Gels with
-gels,
IEF on 13 cm 3-10 NL pre-cast focusing strips (GE Healthcare Bio-sciences)
was performed on an IPGphor unit (GE Healthcare Bio-sciences). IEF
conditions used were an initial gradient up to 500 V for 10 minutes,
a second gradient up to 4000 V for 150 minutes, and a final step at
8000 V for 130 minutes. Following IEF, equilibration was performed
in two steps using an equilibration buffer (EQ) containing 6 M urea,
50 mM Tris-HCl, 30% glycerol and 2% SDS. Step one was performed
in EQ containing 1.5% DTT for 12 minutes and step two in EQ containing
4.5% iodoacetamide for 5 minutes. The second dimension was run on
a 10% SDS-polyacrylamide gel. 2-DE gels were stained with Sypro Ruby
dye (Bio-Rad) according to the manufacturer's recommendations. These
gels were imaged either with UV light as an excitation source and
a 670 nm emission filter (35 nm wide band) on a Kodak image station
2000R or with a 457 nm excitation laser source and a 610 nm emission
filter on a Typhoon imager 9400 (GE Healthcare Bio-sciences AB,
Uppsala,
Sweden). Gels with 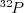 -labeled proteins were
dried in cellophane
wrapping, thereafter exposed to a phosphoimaging plate subsequently
scanned in a Fuji BAS-2500 phosphoimager (Fuji Photo Film Co., Ltd.).
-labeled proteins were
dried in cellophane
wrapping, thereafter exposed to a phosphoimaging plate subsequently
scanned in a Fuji BAS-2500 phosphoimager (Fuji Photo Film Co., Ltd.).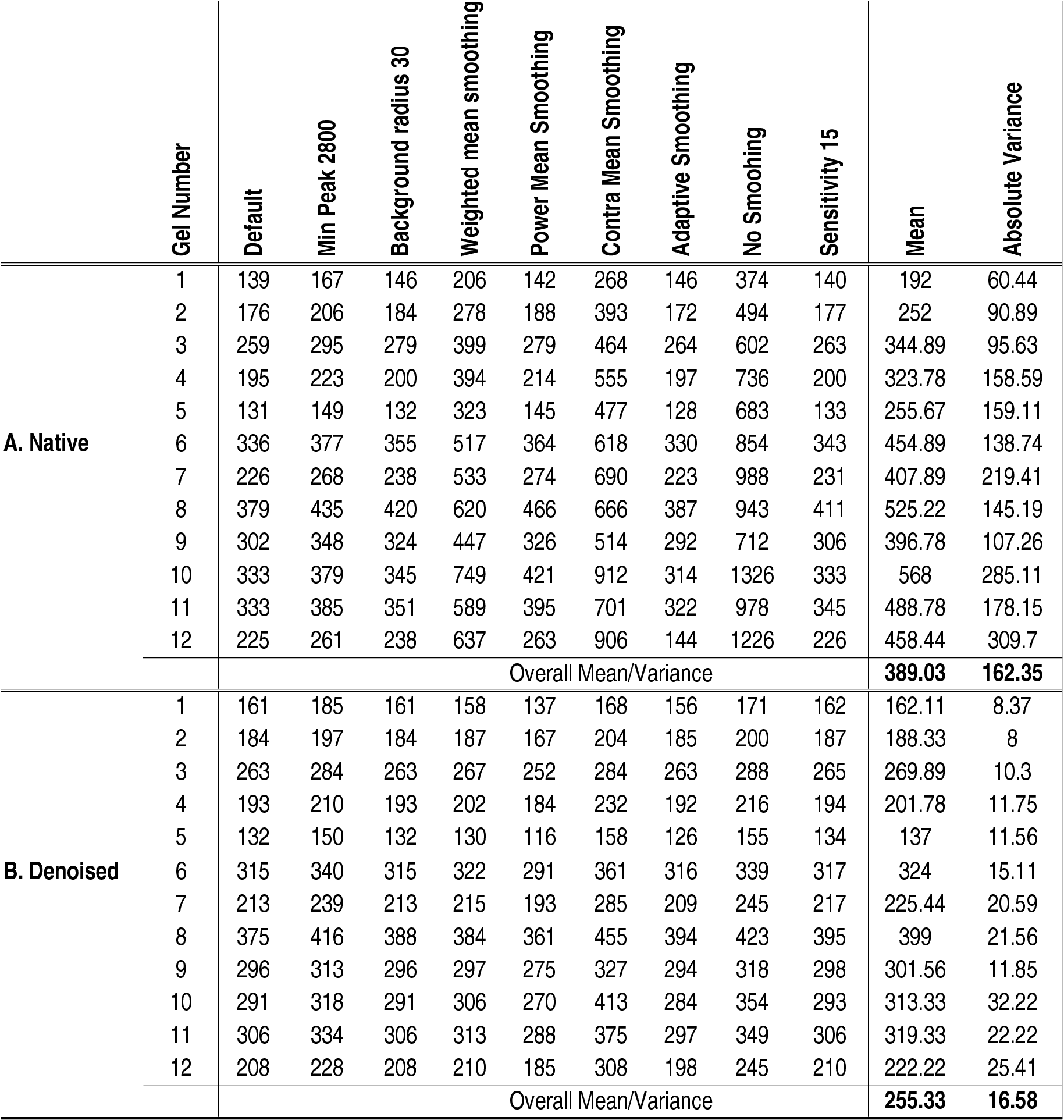 Table 1: Detected spots in native and denoised images of
12 Sypro Ruby stained gels using 9 different spot detection methods.
Section A documents the number of spots detected before denoising.
Section B documents the number of spots detected after denoising.
Because denoising alters spot amplitudes, the number of detected spots
lowers (to 76%). The variance measure reveals that denoising decreases
the impact tuning of spot detection parameters has on the number of
detected spots. See Material and Methods for details on the parameters
used.
Table 1: Detected spots in native and denoised images of
12 Sypro Ruby stained gels using 9 different spot detection methods.
Section A documents the number of spots detected before denoising.
Section B documents the number of spots detected after denoising.
Because denoising alters spot amplitudes, the number of detected spots
lowers (to 76%). The variance measure reveals that denoising decreases
the impact tuning of spot detection parameters has on the number of
detected spots. See Material and Methods for details on the parameters
used.
|
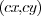 is the center position,
is the center position, 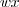 and
and 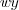 are respectively the width and height and
are respectively the width and height and  is the amplitude
of the dummy spot. Based on this Gaussian bump we made different
simulation
images (Fig. 1).
is the amplitude
of the dummy spot. Based on this Gaussian bump we made different
simulation
images (Fig. 1).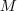 and
and 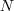 was
obtained by finding the translation of image
was
obtained by finding the translation of image 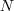 which
maximized
which
maximized
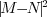 (the
(the 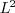 -norm).
This position was found in the cross
correlation image, defined as
-norm).
This position was found in the cross
correlation image, defined as
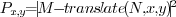 , which
was calculated as
, which
was calculated as
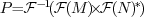 .
* is conjugation.
.
* is conjugation.  and
and
 are respectively
forward and backward Fourier transforms. For more information on cross
correlation see Gonzalez and Woods [[10]]. Wrap around
effects have been avoided by extending both images with a mean padded
border. The image acquisition region covered by the camera did not
allow shifts larger than 10% of the image size, therefore we searched
for the best cross-correlation only within a limited range.
are respectively
forward and backward Fourier transforms. For more information on cross
correlation see Gonzalez and Woods [[10]]. Wrap around
effects have been avoided by extending both images with a mean padded
border. The image acquisition region covered by the camera did not
allow shifts larger than 10% of the image size, therefore we searched
for the best cross-correlation only within a limited range.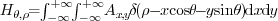 in which
in which 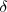 is
the Dirac delta function [[16]],
is
the Dirac delta function [[16]],
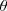 is the angle of the investigated line and
is the angle of the investigated line and 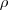 is the
distance from that line to the origin. To increase the sensitivity
of the algorithm, histogram equalization has been performed using
4 bins. Gonzales and Woods [[10]] contains more
information on Hough transforms.
is the
distance from that line to the origin. To increase the sensitivity
of the algorithm, histogram equalization has been performed using
4 bins. Gonzales and Woods [[10]] contains more
information on Hough transforms.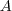 denotes an image, which is
an element of
denotes an image, which is
an element of 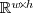 ,
where
,
where 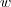 and
and 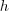 are the
width and height of the image. Pixel positions are written using
subscript.
E.g:
are the
width and height of the image. Pixel positions are written using
subscript.
E.g: 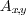 refers to the gray value of the
pixel at position
refers to the gray value of the
pixel at position
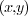 in image
in image 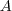 . Adhering to common
interpretation, a gray
value of 0 is black and a gray value of 1 is white. The algorithm
requires that gel images have black spots on a white background: where
the protein expression is large, the gray-value is small. Preliminary
image inversion might be necessary to fulfill this requirement (step
. Adhering to common
interpretation, a gray
value of 0 is black and a gray value of 1 is white. The algorithm
requires that gel images have black spots on a white background: where
the protein expression is large, the gray-value is small. Preliminary
image inversion might be necessary to fulfill this requirement (step
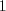 in Fig. 2).
in Fig. 2).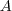 is the
native input
image (Fig. 2B), then the smoothed image
is the
native input
image (Fig. 2B), then the smoothed image 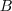 (Fig.
2C) is defined as
(Fig.
2C) is defined as
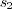 parametrizes
the window size of the filter, which determines the accuracy by which
the background variation will be estimated. The actual filter window
parametrizes
the window size of the filter, which determines the accuracy by which
the background variation will be estimated. The actual filter window
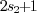 should be larger than the size of the spots we want
to
retain. This filter has zero-phase response, which means that spots
will not be shifted over the surface.
should be larger than the size of the spots we want
to
retain. This filter has zero-phase response, which means that spots
will not be shifted over the surface.
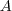 by the
background
by the
background 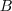 creates the relative strength image (Fig. 2D).
creates the relative strength image (Fig. 2D).
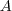 ) than
denominator
(
) than
denominator
(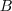 ), resulting in values below 1. Similarly, areas
with less expression
than the neighboring background will have pixel values larger than
1.
), resulting in values below 1. Similarly, areas
with less expression
than the neighboring background will have pixel values larger than
1.![$[0:c]$](img46.png) (Fig. 2E).
(Fig. 2E).
 describes the threshold value which defines the
maximum allowed
image intensity. All positions larger than
describes the threshold value which defines the
maximum allowed
image intensity. All positions larger than  will be set
to
will be set
to  .
In normal conditions
.
In normal conditions  should be
should be  . Eq. 4 formalizes
thresholding.
. Eq. 4 formalizes
thresholding.
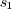 (Fig. 2F),
defined as
(Fig. 2F),
defined as
A median filter will remove all features that are smaller than half
of the window size. Therefore 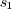 should be much smaller
than
the size of the spot size we want to retain.
should be much smaller
than
the size of the spot size we want to retain.
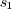 parametrizes the
area-size of
the features to remove, the integer
parametrizes the
area-size of
the features to remove, the integer 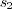 parametrizes the area-size
of the features to retain. The optional floating point number
parametrizes the area-size
of the features to retain. The optional floating point number  parametrizes the threshold (Eq. 4), which
by default
is set to 1.0. Under normal circumstances this is a good choice.
Presence
of much white noise might require a decreased
parametrizes the threshold (Eq. 4), which
by default
is set to 1.0. Under normal circumstances this is a good choice.
Presence
of much white noise might require a decreased  to e.g.,
0.9. The
optional white_spots boolean can be set
when the
image has white spots on a black background. It will then automatically
invert the image before and after the actual denoising. The software
can be used online at
http://analysis.yellowcouch.org/2ddenoising.html
to e.g.,
0.9. The
optional white_spots boolean can be set
when the
image has white spots on a black background. It will then automatically
invert the image before and after the actual denoising. The software
can be used online at
http://analysis.yellowcouch.org/2ddenoising.htmlFUNCTION denoise, image, s1, s2, c = 1.0, white_spots = 0
if white_spots then image = max(image) - image ; invert if necessary
bg = median(image,s2) ; calculate the background variations
im = image / bg ; remove them through division
im <= c ; select only areas darker than the environment
im >= 0 ; no -inf number when dividing by -0
im = median(im,s1) ; remove details and return
if white_spots then im = max(im)-im ; back inversion
return, im
Results and Discussion
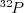 labeled
phosphoprotein. Spots
below the visual threshold in the native image become more visible
and have more distinct positioning after denoising. The first
immuno-blot
demonstrates sharpening of spots. The Sypro Ruby gel shows how
artefacts
introduced through air under the gel are removed as well. The very
dark spots in the denoised
labeled
phosphoprotein. Spots
below the visual threshold in the native image become more visible
and have more distinct positioning after denoising. The first
immuno-blot
demonstrates sharpening of spots. The Sypro Ruby gel shows how
artefacts
introduced through air under the gel are removed as well. The very
dark spots in the denoised 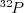 phosphoprotein
labeled gel are
places where the phosphoimaging plate has been over-saturated by
radiation.
Denoising can help researchers in observing spots that are relevant
but below their visual threshold.
phosphoprotein
labeled gel are
places where the phosphoimaging plate has been over-saturated by
radiation.
Denoising can help researchers in observing spots that are relevant
but below their visual threshold.Figure 3 - Illustration of gel denoising -Various
examples of the contrast enhancement process on Sypro Ruby, gels of
$^{32}P$ labeled phosphoproteins and immuno-blots. Note the high
background in the 32P phosphoprotein detection image compared to Sypro
Ruby.
| Technique |
Native |
Denoised
|
|
| Sypro Ruby | 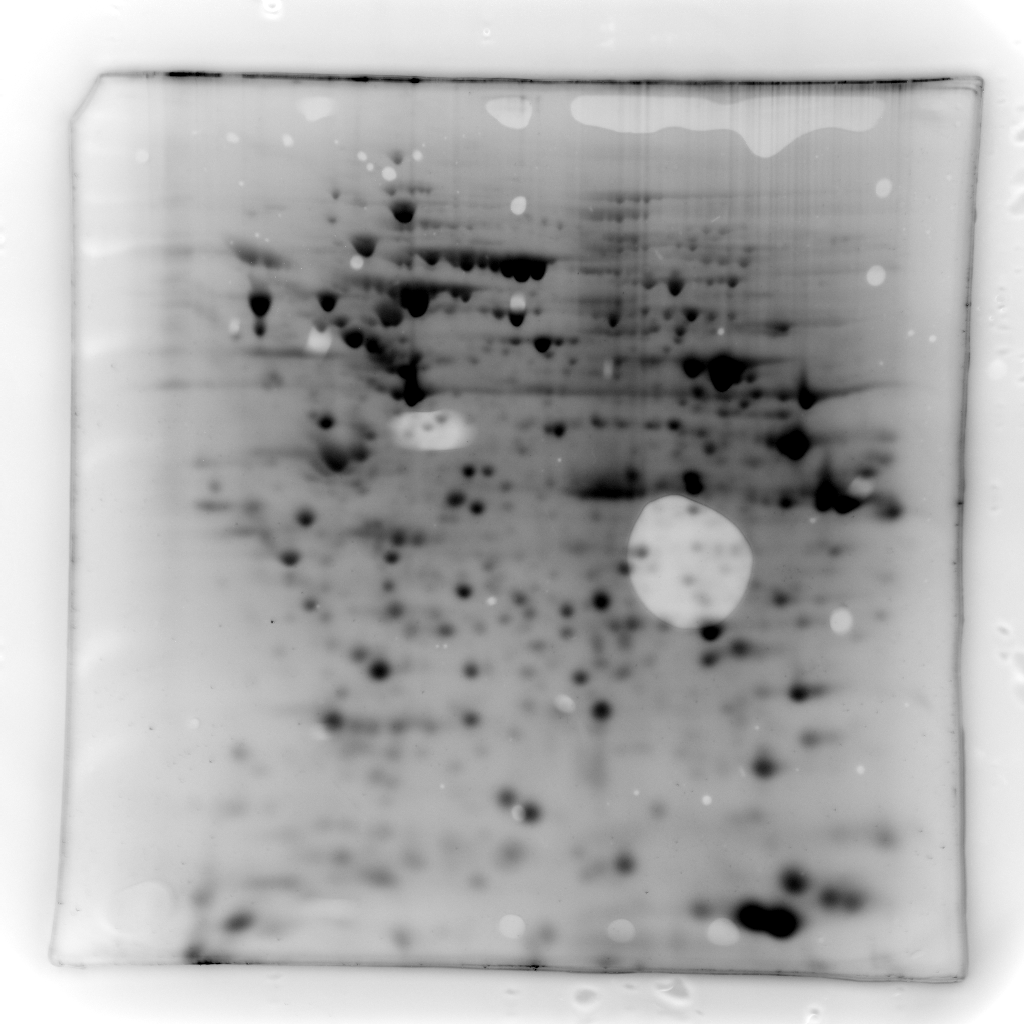 |
 |
|
| 32P phosphoprotein
detection by phosphoscreen imaging |
 |
 |
|
| Immuno-blot with
background variations |
 |
 |
|
| Immuno-blot with low
background variations |
 |
 |
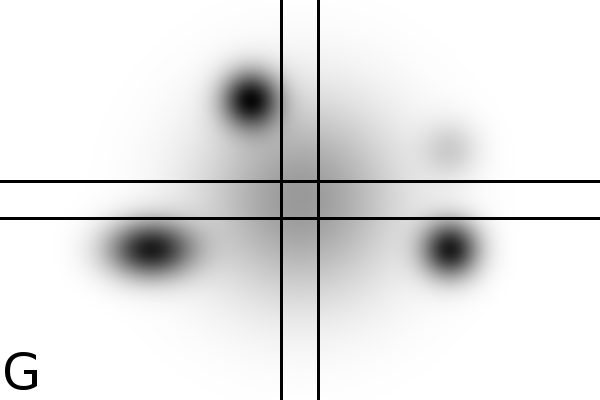
Figure
4 - Denoising Improves Automatic Orientation of 2DE Images - Two examples on how
denoising helps in determining the orientation of a gel. Without
denoising the background variations might influence the measured
orientation. The black lines shows the measured direction(s) on the
different images using a Hough transform (see Material and Methods).
Equally optimal directions are all shown, possibly resulting in
multiple directions for the same image. The left column shows two
original images (A, C). The right column shows their denoised versions
(B, D).
 |
 |
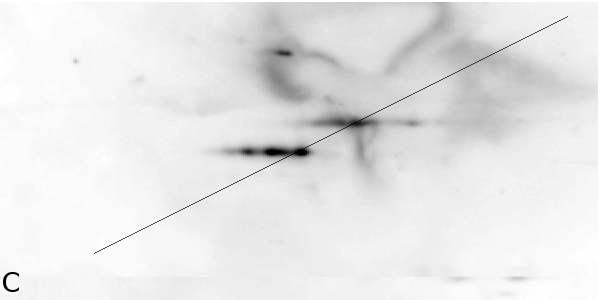 |
 |
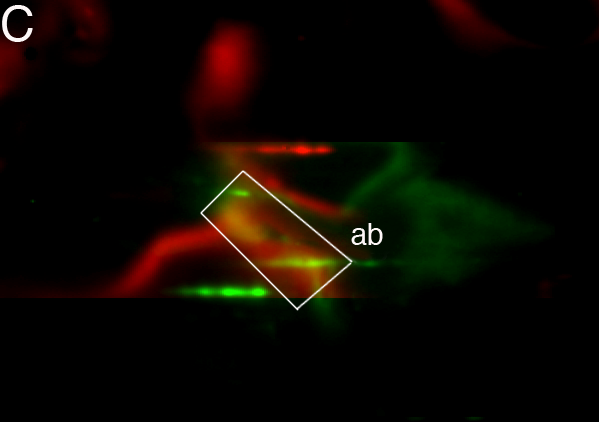
Image alignment - Alignment is necessary to
compare two 2-DE
gel images with a slightly different information content. Spot position
comparison can reveal important differences between biological samples.
Therefore we also relied on 2-DE gel images from slightly different
biological systems. After manual alignment and orientation of 73 2-DE
gel images (based on 2 known spots) we randomly selected 1050 image
pairs. For every pair we calculated the overlay alignment as described
in Material and Methods. An alignment was considered to be a hit when
it deviated no more than 1 mm (or 12 pixels at 300 DPI) from the manual
alignment. Without denoising, the alignment procedure performed correct
in 48% of the cases, while denoising increased the hit rate to 61%.
In the native images, the error (or misalignment) has a standard
deviation
of 25.7 pixels while this decreased to 18.59 pixels using the denoised
images. Fig. 5 shows how the presence of
strong
background variations might influence overlay alignment. Supplementary
Table 'align.xls' contains all couples.
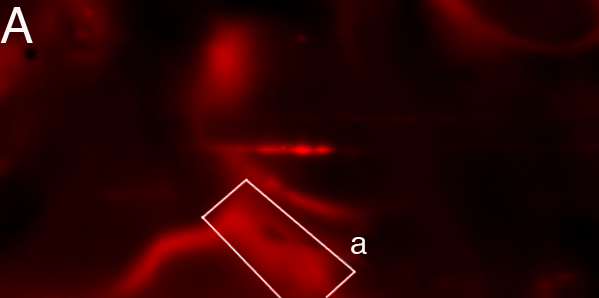 |
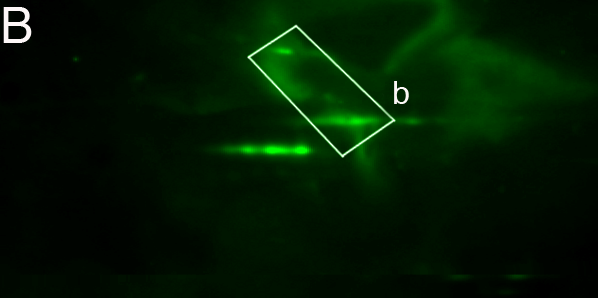 |
 |
 |
Signal-to-background ratio - The signal-to-background ratio was calculated by dividing the gel maximum intensities by the gel mean intensities. The averaged signal-to-background ratio was 4.76 for non denoised images and 147.83 for denoised imaged, indicating an increase in signal level by a factor 31. Assessment of background stability has been performed by calculating the normalized standard deviation. Without denoising the RMS was 0.1, while after denoising this became 0.06. In our experiments, denoising removed 36% of the signal variance, while still improving the analysis properties of the 2-DE gel images. Supplementary Table 'energy.xls' contains the detailed information.
 |
 |
 |
 |
 |
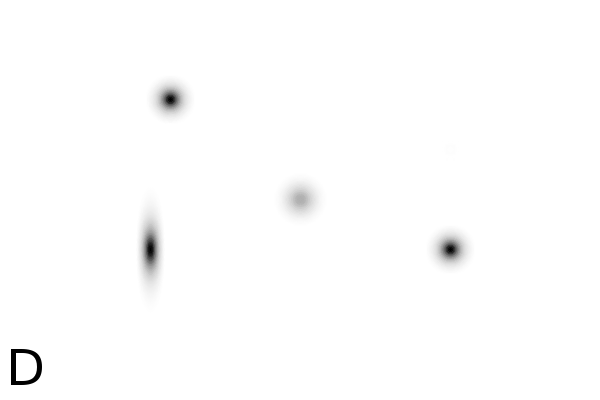 |
 |
 |
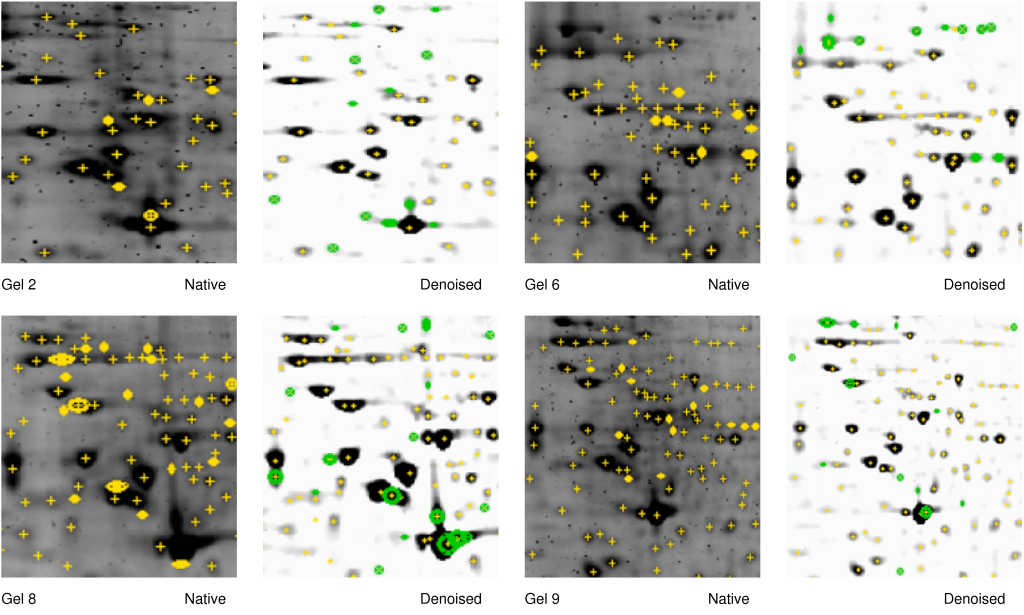 Figure 6 - Denoising improves spot detection in darkly stained areas - Gel
numbers 2,6,8 and 9 refer to Table 1. The denoised image is shown on
the right side of the 2-DE image. The same spot detection parameters
were used in all cases (see Material and Methods). The denoiser ran
with s1=3 and s2=23. Spots marked by yellow circles are only detected
in native images. Spots marked with green circles are detected only in
denoised images. Note that closely located intensely stained spots
(Gels 2, 8, lower right) and regions with low signal-to-strength ratio
(Gels 2, 6, 8, 9, upper part) are resolved after denoising.
Figure 6 - Denoising improves spot detection in darkly stained areas - Gel
numbers 2,6,8 and 9 refer to Table 1. The denoised image is shown on
the right side of the 2-DE image. The same spot detection parameters
were used in all cases (see Material and Methods). The denoiser ran
with s1=3 and s2=23. Spots marked by yellow circles are only detected
in native images. Spots marked with green circles are detected only in
denoised images. Note that closely located intensely stained spots
(Gels 2, 8, lower right) and regions with low signal-to-strength ratio
(Gels 2, 6, 8, 9, upper part) are resolved after denoising.
|
Tandem setup for quantitative analysis - Regarding quantitative analysis, e.g, to assess protein regulation, or measure protein expression, one can combining the spots detected in native and denoised images to create an improved master image (scout image). Subsequent spot quantification on the native gels, will provide the researcher with the required protein expression information. However, one must be careful to quantitatively interpret gels with high background variation. Background variations can be part of the investigated biological system or be introduced through technical variations in the experiment. Preferably, the experiments should be replicated to avoid misinterpretation [[11]].
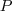 )
as
a combination of signal (
)
as
a combination of signal (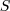 ), multiplicative
background (
), multiplicative
background (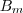 )
and additive background (
)
and additive background (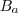 ).
).
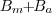 . Subtraction of this background
leads to a denoised
image
. Subtraction of this background
leads to a denoised
image 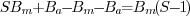 ,
which still has the
full impact of the multiplicative noise. Division of the signal by
its background
,
which still has the
full impact of the multiplicative noise. Division of the signal by
its background
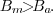 This fact, combined with our empirical observation, leads us to
conclude
that background variations are mainly multiplicative.
This fact, combined with our empirical observation, leads us to
conclude
that background variations are mainly multiplicative.![$[0:1]$](img59.png) , while the background
will
map onto
, while the background
will
map onto 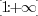 . If we would work with white spots
on a black
background the region of interest would become
. If we would work with white spots
on a black
background the region of interest would become 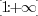 ,
which
is difficult to normalize. The discrepancy between B/W and W/B images
occurs because we have different relations:
,
which
is difficult to normalize. The discrepancy between B/W and W/B images
occurs because we have different relations: 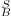 and
and 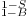 ,
with
,
with 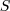 being the real signal and
being the real signal and 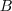 being the
measured background.
See Supplementary Figures for the result of different rescaling methods.
being the
measured background.
See Supplementary Figures for the result of different rescaling methods.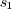 ,
, 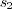 and
and  ,
the algorithm performs
linearly to the size of the image. All steps in the algorithm:
smoothing
(Eq. 2), division (Eq. 3), thresholding
(Eq. 4) and median filtering (Eq. 5)
can be performed in
,
the algorithm performs
linearly to the size of the image. All steps in the algorithm:
smoothing
(Eq. 2), division (Eq. 3), thresholding
(Eq. 4) and median filtering (Eq. 5)
can be performed in 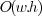 , with
, with 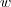 and
and 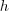 , respectively the
width and height of the image.
, respectively the
width and height of the image.Concluding Remarks
We presented a denoising algorithm that requires no user input and provides an execution time linear to the image size. The algorithm relies on division of the original image by the estimated background variation and therefore only works on images with black spots on a white background. The algorithm was particular effective in removing background variance from immunoblots and radioisotopic labeled gels. Denoising improved the working of algorithms for gel orientation and overlay alignment. Spot detection on denoised Sypro Ruby images (using standard software) confirmed that the algorithm improves spot detection in areas with high background variation and in areas with closely located high-intensity spots. Because the algorithm modifies spot intensities, denoised images cannot be used for spot quantification. A future application for the algorithm could be the generation of a scout image to guide automatic spot recognition algorithms and quantify spot volumes in the original image, eliminating the manual pre- analysis processing of 2-DE images.
Acknowledgments
Bibliography
| 1. | Recent Advances in gel-based proteome profiling techniques Y. Hu, X. Huang, G.Y. Chen, S.Q. Yao Molecular Biotechnology, volume 28, number 1, pages 63-76, September 2004 |
| 2. | Proteomics in acute myelogenous leukemia (AML): methodological strategies and identification of protein targets for novel antileukemic therapy. Gry Sjoholt, Nina Ånensen, L Wergeland, E McCormak, Ø Bruserud, Bjørn Tore Gjertsen Current Drug Targets; volume: 6; number: 6; pages: 631-646; 2005 |
| 3. | High Resolution two-dimensional electrophoresis of proteins P. H. O'Farrell J. Biol. Chem.; volume: 250; number: 10; pages: 4007-21; May 25; 1975 |
| 4. | Identification of extracellular and intracellular signaling components of the mammary adipose tissue and its interstitial fluid in high risk breast cancer patients: toward dissecting the molecular circuitry of epithelial-adipocyte stromal cell interactions J.E. Celis, J.M. Moreira, T. Cabezon, P. Gromob, R. Friis, F. Rank, I. Gromova Mol Cell Proteomics; volume: 4; number: 4; pages: 492-522; February; 2005 |
| 5. | Current two-dimensional electrophoresis technology for proteomics. A. Gorg, W. Weiss, M.J. Dunn Proteomics; volume: 4; number: 12; pages: 3665-3685; Dec; 2004 |
| 6. | Analysis of two-dimensional electrophoresis gels K. Conradsen, J. Pedersen Biometrics; volume: 48; pages: 1273-1287; 1992 |
| 7. | Analyzing Two-Dimensional Gel Images Roy Anindya, R. Lee Kwan, Hang Yaming, Mark Marten, Raman Babu institution: Department of Mathematics and Statistics, University of Maryland; August; 2003 |
| 8. | Statistical methods for proteomics F. Seillier-Moiseiwitsch, D.C. Trost, J. Moiseiwitsch In proceedings of Methods in Molecular Biology. Humana Press Inc, NJ. Volume 184, 2002 |
| 9. | Denoising method and apparatus Maeton-dong, Yeontong Gu, Suwon Si, Gyeonggi Do Patent application nr PCT/KR2004/002428; International publication number WO2005/032122 A1, April 2005 |
| 10. | Digital Image Processing Rafael C. Gonzalez, Richard E. Woods Prentice Hall; chapter: 7; pages: 432-438; address: Upper Saddle River, New Jersey 07458; edition: 2nd; 2002 |
| 11. | Towards validating a method for two-dimensional electrophoresis/silver staining Wolfgang Schlags, Michael Walther, Mohammed Masree, Martin Kratzel, Christian R. Noe, Bodo Lachmann Electrophoresis; volume: 26; pages: 2461-2469; 2005 |
| 12. | Single Cell profiling of potentiated phospho-protein networks in cancer cells J.M. Irish, R. Hovland, P.O. Krutzik, O.D. Perez, Ø. Bruserud, B.T. Gjertsen, G.P. Nolan Cell; volume: 118; pages: 217-228; 2004 |
| 13. | Analysis of acute myelogenous leukemia: preparation of samples for genomic and proteomic analysis Bjørn Tore Gjertsen, A.M. Oyan, B. Marzolf, Randi Hovland, G. Gausdal, Stein Ove Doskeland, K. Dimitrov, A. Golden, K.H. Kalland, L. Hood, Ø. Bruserud J Hematother Stem Cell Res; volume: 11; number: 3; pages: 469-81; June; 2002 |
| 14. | Novel (Rp)-cAMPS analogs as tools for inhibition of cAMP-kinase in cell culture. Basal cAMP-kinase activity modulates interleukin-1-beta action B. T. Gjertsen, G. Mellgren, A. Otten, E. Maronde, H. G. Genieser, B. Jastroff, O.K. Vintermyr, G. S. McKnight, S. O. Doskeland J Biol Chem; volume: 270; number: 35; pages: 20599-607; Sep 1; 1995 |
| 15. | Methods and Means for Recognizing Complex Patterns Hough, P.V.C US Patent 3,069,654; 1962 |
| 16. | The Fourier Transform and its applications R. Bracewell Chapter 5. The Impulse Symbol; New York: McGraw-Hill; pages 69-97, 3th edition; 1999 |
| 17. | IDL, The Interactive Data Language, v6.1 |
| 18. | The Art of Computer Programming Donald Knuth Addison-Wesley; chapter: 1.2.11: Asymptotic Representations; pages: 107-123; volume: 1; edition: 3th; 1997 |
| http://werner.yellowcouch.org/ werner@yellowcouch.org |  |
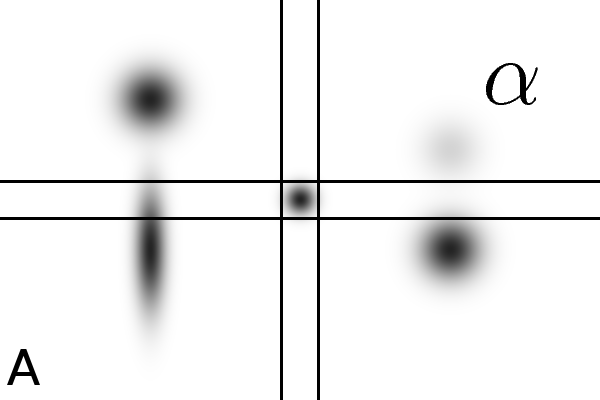
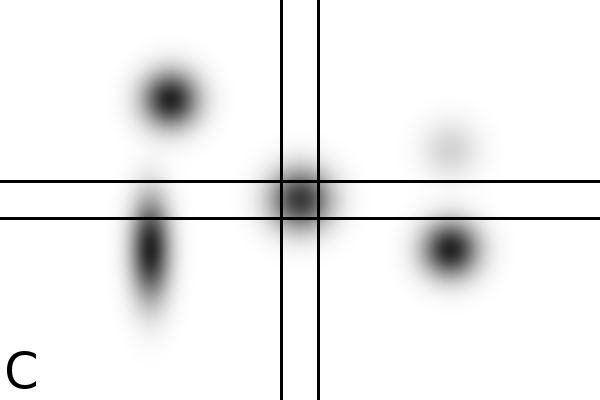
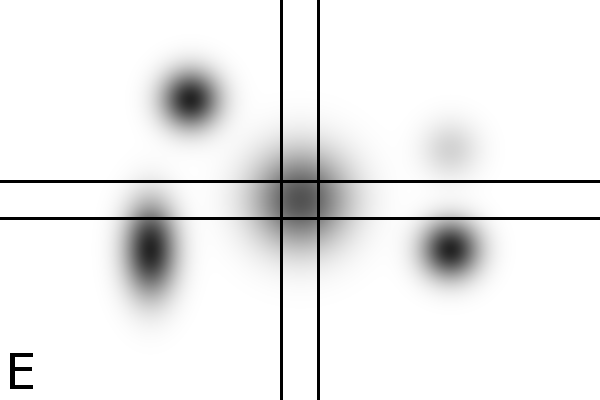
 Figure 2: Image denoising flowchart
- The denoising process estimates the background intensities (step 2)
and uses them to obtain the relative protein expression (step 3). All
non relevant information (protein expression lower than the estimated
background) is removed through thresholding (step 4) and median
filtering (step 5). Input images should have black spots on a white
background. If the original images have white spots on a black
background (A) they should be inverted (step 1 and 6), see Results and
Discussion section. Image denoising is useful for many applications,
including visualization, gel orientation, gel alignment and spot.
Figure 2: Image denoising flowchart
- The denoising process estimates the background intensities (step 2)
and uses them to obtain the relative protein expression (step 3). All
non relevant information (protein expression lower than the estimated
background) is removed through thresholding (step 4) and median
filtering (step 5). Input images should have black spots on a white
background. If the original images have white spots on a black
background (A) they should be inverted (step 1 and 6), see Results and
Discussion section. Image denoising is useful for many applications,
including visualization, gel orientation, gel alignment and spot.