| Home | Papers | Reports | Projects | Code Fragments | Dissertations | Presentations | Posters | Proposals | Lectures given | Course notes |
MarFlow Sample TrackingWerner Van Belle1* - werner@yellowcouch.org, werner.van.belle@gmail.com Abstract : Tracking samples throughout many different organisations which all use different labeling protocols and are located at different geographical locations is difficult. This R&D program aims at the creation of a sample tracking system. The system will offer a) a labelling protocol designed for storage and unique identification of samples integrating many different labeling techniques b) a decentralised storage capacity (every organisation can store the data locally) and c) a security model which will take into account the ethics of sample exchange. A second aspect of the project aims at offering integration and data exchange towards existing sample tracking systems and analysis programs. We believe that this project and associated effort will reduce the costs of future information systems as well as increase cooperation between different research groups. The longer term goal of this project is the development of one or more commercial products related to sample tracking in cooperation with interested partners.
Keywords:
peer to peer, distributed systems, ambient intelligence, ubiquoutous computing, pervasive actors |
1 Introduction
In Norway, biobanks and high throughput experiments are driven by advanced experimental techniques such as micro-arrays, mass spectrometry, HPLC, X Ray crystallography and others. These techniques can be used to screen for specific activities, to develop detailed models of biological systems or to obtain detailed understanding of single samples. Such experiments typically consist of a pipeline of different techniques, in which samples are transferred between different stages. In every stage a sample is analysed and then, depending on the outcome, routed in the pipeline. Given the huge amount of samples transferred in these pipelines, research groups struggle with sample tracking. Some users (notable hospitals) make use of locally developed sample tracking systems, some use off-the-shelf software without external inputs or outputs, while others use pen and paper. Sometimes, other ingenious techniques are appropriate such as RF identifaction and DNA markers, which might be a much more natural way of identification.
In this context, where integration of different labeling protocols is necessary and where sample exchange between different research groups require a decentralised approach, many laboratory information management systems do not scale up.
In this project we propose the creation of a system that will help in tracking samples, thereby integrate existing sample tracking techniques. For groups currently in need of a sample tracking system, SampleTrack will a) offer tracking using barcode labelling and b) offer a flexible local storage peer to peer information system. For groups who already make use of a sample tracking system, SampleTrack will a) integrate with the existing system and b) integrate with the local labelling protocol. A large emphasis will be placed on offering a flexible access right system to guarantee privacy and honour ethics. E.g; sending out local patient information should not be done, however it should still be possible to track the sample over different organisations.
2 Requirements / Goals
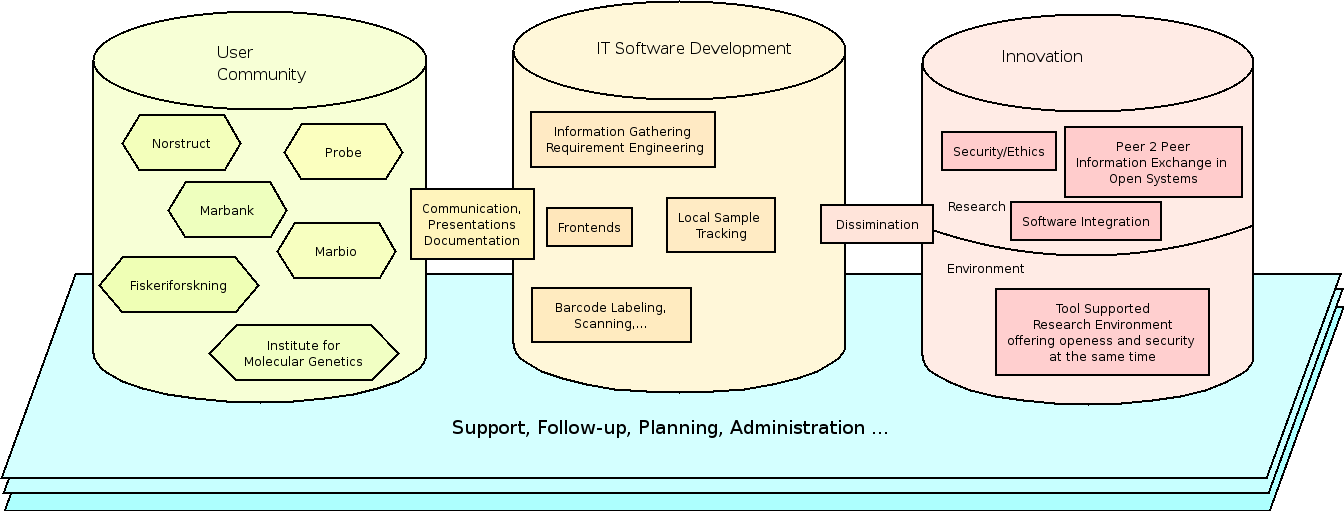
The requirements for a sample tracking systems that spans multiple organisations are different from standard in-house sample tracking systems. We have been in communication with a number of different research groups. Among them are Marbank (the national marine biobank), Marbio (the analysing laboratory associated with Marbank), the molecular genetics group at the University of Tromsø, Norstruct (the Norwegian structural biology centre), Labforum (the micro-array facility at the University of Tromsø) and Probe (the proteomics unit at the University of Bergen). This lead to the identification of a number of requirements for a sample tracking system. We describe them briefly below.
2.1 R1: Unique identification
The first requirement of any sample tracking system is that all samples can be uniquely identified. At any moment it should be possible to look at a sample and retrieve the associated sample information. Depending on the research group the requirements are slightly different.
- R1.1: Sample Labelling: Some research groups do not yet have a suitable sample labelling protocol. For these research groups it is necessary to offer a well designed standard.
- R1.2: High throughput: In order to be viable in the near future, the labelling protocol should be able to handle large quantities of samples. This means that it should be possible to quickly label a lot of samples. This can be done by making use of barcode labelling and by group labelling. E.g; instead of labelling every vial in a rack, it is also possible to refer to a position in a uniquely labelled rack.
- R1.3: Integration: Some research groups already have a sample labelling strategy. It should be possible to use these local labelling protocols interwoven with the labelling protocol we will develop. Especially our own expertise with DNA markers and RFC identification will bring a new input into the aspects of laboratory information management systems.
2.2 R2: Flexibility
The system should be flexible to use. This means that the steps between installing the software and actually using it should be as small as possible.
- R2.1: Associated Information. Every research group has its own information it wants to associate with a sample. Some research groups want to associate physical location with a sample, others want to know to which project a certain sample belongs. It is difficult to foresee what information is necessary, therefore, it is essential that the system is extremely flexible in adding new information to samples.
- R2.2: Process Information. Sometimes samples are tracked throughout a process. It should be possible to follow a sample within a certain process, retrieve the sample history and so on.
- R2.3: User Interface. The system should offer a number of easy to use front ends and installers.
- R2.4: Sample Searching: The system needs to offer a way to search for specific samples, based on their associated meta-information.
2.3 R3: Decentralised
One of the key requirements that came forth by a number of research groups is that a centralised sample tracking system would be highly inappropriate. Most research groups want to have local control over their own data.
- R3.1: Decentralised. The system should be decentralised, which means that all sample informations of local created samples should be stored locally.
- R3.2: Distributed. Although all sample information is stored locally, it is still necessary that information can be shared between multiple partners. For instance, when a sample is transferred from one research group to another, the exchange of visible information should be transparent.
- R3.3: Disconnected. In some situations samples are handled at remote locations, where there is no immediate connection towards the sample tracking system. In such a disconnected setup, it is necessary that the system synchronises data transparently and smoothly.
- R3.4: Redundancy: To ensure that data is not lost, it is necessary to have some redundancy in the sample tracking system.
2.4 R4: Integration
Currently in some companies, sample tracking is performed by using all kinds of tools. In order for this project to succeed it is crucial that SampleTrack will be able to integrate with other systems.
- R4.1: Import/Export: depending on the system already in use it might be necessary to import and export data in and out of the system. E.g: export data to excel files or input data from excel files.
- R4.2: Synchronous Linking: when possible it might be helpful to create an immediate link towards already existing infrastructure.
- R4.3: Extensibility: the sample tracking system we will develop should be accessible by other programs as well. This will ensure that research groups can alter and modify the system as they need.
2.5 R5: Ethics
Another big concern of the different research groups was that the system should support ethics and allow to associate private information with samples.
- R5.1: Security: Given the ethical committee its decisions about the visibility of patient information, it is necessary to ensure that suitable security policies can be linked to the samples. E.g; patient data should not be visible by anybody else beside the M.D. himself.
- R5.2: Access Domains: Sometimes access should be granted towards anybody working within the research unit or within a specific research project.
3 Related Work
There exists many laboratory information systems, which are typically very good at satisfying local requirements for one research lab, however they almost never succeed in allowing the exchange of samples (and associated information) towards other systems and research groups. Also, they fail to integrate more advanced techniques of sample identification, such as DNA markers. Furthermore, communication with vendors of commercial sample tracking systems has shown that information exchange is often considered to be a bad business strategy. Commercial systems are closed, have an expensive and unnatural licensing and software tailoring requires support from the original seller. Clearly such a closed world strategy and extensive payments per license are difficult to integrate in a cooperative research environment.
This proposal aims at bringing a usefull sample tracking system towards many reasearch groups, whilst offering a natural integration between them.
4 User Community
In this section we briefly cover some of the requirements we are already aware of with our partners
- Marbank: every sample in the marine biobank will be associated with multimedia (digital photos, video, literature), geographic information (date, geographic position, field preparation, notes, storage in transit, hazard), biological information (species, common name, weight, length, sex, age) and expert annotations (determined by, reference annotations). The preservation of every sample, initial and available amount and location in the freezer (freezer name, section, rack and box) will also be stored. Further useful information is tracking which research groups are actually using which sample. Another aspect of Marbank is that samples will be gathered off-shore, which means that when the floating lab comes back all data should propagate to the appropriate locations.
- Marbio: Marbio will test for specific sample activity by making use of a pipeline. the typical process starts by extracting the preserved sample into a water phase and an organic phase. In this step a sample is transformed into multiple other samples using specific preparation methods. These extracts are then routed through a HPLC fractionation which will split the extracts into fractions. For every fraction we need to keep track of the column, eluation/gradient, time/top, absorbans, flow speed and storage parameters. These different fractions will then be tested for bioactivity using different protocols. These can be tests on anti-tumor, anti-inflammatory, anti-viral and anti-bacterial tests. The sample tracking systems should keep track of all these samples and the processes that have been applied to them.
- Norstruct: The FUGE-technology platform for structural biology is carrying out structural analysis on biologically active macromolecules, both in connection with internal projects in the laboratory and for external partners nation-wide that are using the services we provide. The selected target for structural studies will be carried through a ``pipeline'' including many steps; target selection, evaluation and identification, sequencing, cloning, recombinant expression, biochemical and biophysical characterisation, structure determination, crystallisation, data collection and structure analysis. Each of the steps will, for each target, create a range of data (of variety of formats) that needs to be stored, both for documentation purposes and for subsequent analyses for optimisation of procedures. Since Norstruct is a national laboratory with obligations towards ``external customers'', clear and easy access to data and documentation is required.
Furthermore, there are relations with the following partners:
- Probe: Probe is currently working together with a Belgian unit to develop their local sample tracking system. An immediate integration with their system will make sure that the software forms a bridge between different research groups. At probe samples are analysed using multiple different techniques. For instance for a Maldi TOF analysis it is important to keep track of the digestion procedure followed, which digestive was used, what time, what buffering. Afterwards it is necessary to keep track on which spots the sample was placed and what the output of the machine was given a certain laser strength, polarity and technician. Extra useful information is to know from which lab the sample came and for which project.
- The Institute For Molecular Genetics: (at the university of Tromsø) currently looks for a solution to keep track of plasmids in a freezer. Information associated with every plasmid is the creator, who has used it, how much is left and how much was originally present, the name of the plasmid and the location in the freezer.
- Fiskeriforskning: Norut IT has been working together with Fiskeriforskning for a couple of years already with respect to sample tracking in the area of fish identification using RFC chains and DNA markers, based on microsattelites and SNP's. These projects have been funded by Innovation Norge (through Mabit). Currenlty we believe that the experience we got from there should certainly make it possible to broaden our scope and involve more partners and start developing a more integrated, scaleable and reusable software line.
5 Scientific Program
The overall program we present contains three distinct elements. The first element is the user community which drives the project by using the software and communicating with us with respect to their needs. The second element is the development of usable prototypes in order to satisfy the needs of the different users and the last part is the innovative part. The innovation of this project lies in the application and refinement of techniques which are nowadays being developed in the research community. The SampleTrack program will bring this research to an applied level by a) using research where applicable, b) making research applicable where necessary and c) inventing new techniques where none exists. A second innovation of this project lies in the value it offers to the scientific community. The SampleTrack system offers tool support for the scientific community to work together in a better and easier way. This will especially become important whenever research laboratories will transfer samples in high throughput mode.
6 Work Program
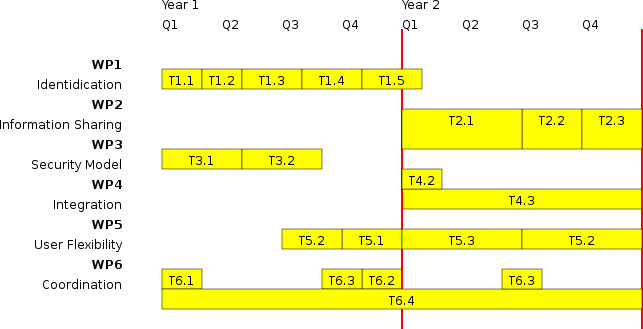
The work program (covering the entire scientific program) consists of 6 work packages, each divided in a number of tasks. Due to space constraints we keep the description as small as possible. The total project contains 56 MM spanning 2 years.
6.1 WP1: Sample Identification & Local Information Storage [11MM]
This work package is concerned with
- T1.1 [1MM]: gather overview of existing labelling protocols in the different groups
- T1.2 [1MM]: designing and identification model, which integrates multiple protocols as one
- T1.3 [3MM]: develop internal meta-data representation of samples and processes, which is secure and allows to adding extra information
- T1.4 [3MM]: develop program and API to store and access data
- T1.5 [3MM]: deploy barcode printer, scanner and driver software
6.2 WP2: Development of Peer to Peer Information Exchange Network [12MM]
- T2.1 [6MM]: develop lab interconnection protocol which will connect different local databases to each other
- T2.2 [3MM]: develop propagation protocol throughout the peer to peer network, which introduces redundancy
- T2.3 [3MM]: develop synchronisation protocol after reconnection
6.3 WP3: Security Model [4MM]
- T3.1 [2MM]: develop security model, using multi-user key-rings and so on, avoiding user identification
- T3.2 [2MM]: develop identification domains
6.4 WP4: Software Integration [10MM]
- T4.1 [9MM]: develop linker programs toward existing databases & tools
- T4.2 [1MM]: develop API useable by other programs, including a web back-end
6.5 WP5: User Flexibility [9MM]
- T5.1 [3MM]: develop installer
- T5.2 [3MM]: develop application front-end
- T5.3 [3MM]: develop web front-end
6.6 WP6: Administration, Communication & Dissemination [10MM]
- T6.1 [1MM]: do requirement analysis
- T6.2 [2MM]: SampleTrack website covering program, documentation and so on
- T6.3 [2MM]: Presentations and seminars about SampleTrack
- T6.4 [10%]: Administration and project management
7 Milestones
The program consists of two major milestones. M1: The first milestone (at the end of the first half year), is the deployment of a local information tracking system at multiple locations. This early milestone will give us feedback throughout the remainder of the project and will allow us to refine the information tracking system further. M2: The second milestone is the opening of the information tracking system towards different organisations. This milestone is located in the middle of the second year. The last milestone refers to the dissemination of the results obtained in this high visibility project.
8 Innovation
Despite the practical nature of this project, there are a number of aspects which are strongly rooted in current day research. Below we briefly touch upon them.
- Regression Analysis: In order to be able to guarantee that any new version of the software is compatible with old versions (backward compatibility) we will investigate the use of Petri-nets as a tool to do automatic regression testing. Also, the applicability of Petri-nets should make it possible to verify whether any future versions will still have possibilities to offer new behaviour (forward compatibility).
- Correct Interface Usages: If software is developed to link with existing tools and data formats, it is crucial to be sure that the format is exactly as it is supposed to be. To guarantee this we will use formal interface and data descriptions which can be checked vigorously. Also, this should make it possible to verify that external programs interfacing with the SampleTrack sample tracking system behave properly.
- Secure peer to peer information exchange. The exchange of information in peer to peer networks is nowadays used to transfer (often illegal) data between partners. The decentralised nature of peer to peer networks makes them very robust but at the same time very difficult to handle. How this can be done properly is still one of the areas of investigation. Especially the subproblems of identification of users and samples as well as the honouring of security requirements is still a research challenge.
- Automatic data synchronisation. Peer to peer networks rely upon some form of automatic data synchronisation. This field, which is closely related to concurrent systems, requires solutions for every specific problem at hand. In this specific case of sample tracking it is necessary to have some information exchange which follows the actual physical exchange of samples. Nevertheless at the moments when multiple data fields exists, it is necessary to have some intelligent synchronisation mechanism.
- The development of a security model is highly dependent on how users think about their data. There is little knowledge on how biological and ethical sensitive information propagates throughout human networks and where information naturally stops. Knowing this information is crucial to develop a highly suitable security model. Such a model should allow us to develop a trust propagation model. In the context of secure peer to peer networks, Norut IT has requested for funding at Forskningsrådet to investigate the possibilities of creating a scalable and secure peer to peer network. This project is called TRUSTNET.
| http://werner.yellowcouch.org/ werner@yellowcouch.org |  |